
Dynamic Interactions of Fully Glycosylated SARS-CoV-2 Spike Protein with Various Antibodies | Journal of Chemical Theory and Computation

Accurate 3D Characterization of Catalytic Bodies Surface by Scanning Electron Microscopy - Gontard - 2019 - ChemCatChem - Wiley Online Library

Atomistic Simulations and Network-Based Energetic Profiling of Binding and Allostery in the SARS-CoV-2 Spike Omicron BA.1, BA.1.1, BA.2 and BA.3 Subvariant Complexes with the Host Receptor: Revealing Hidden Functional Roles of the
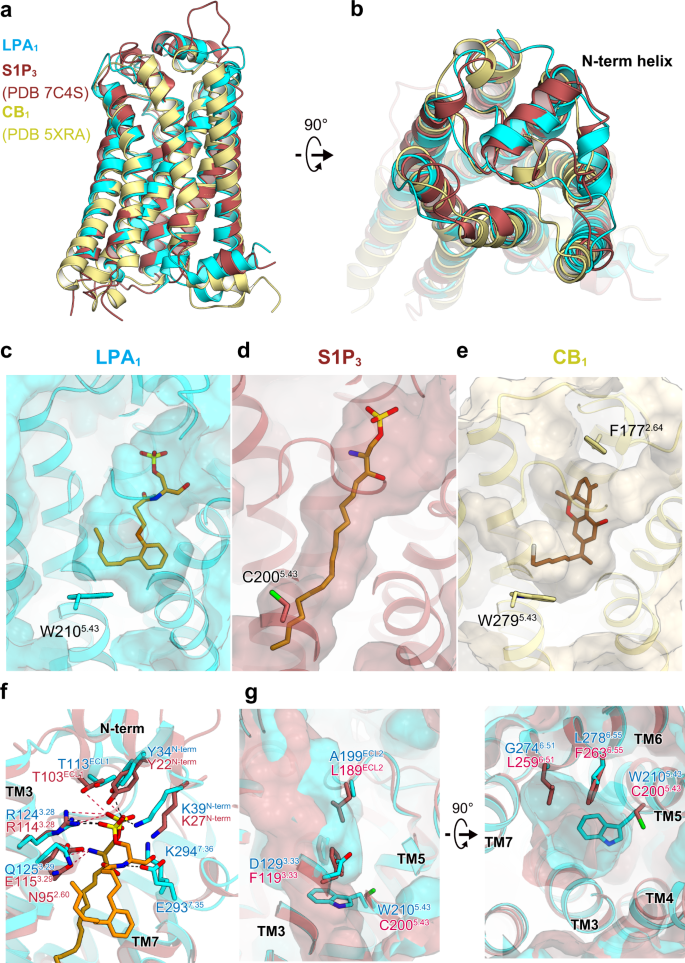
Structure of the active Gi-coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist | Nature Communications

Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C6‐Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C6‐Methyltransferase - Tsutsumi - 2022 - Angewandte Chemie International Edition - Wiley Online ...
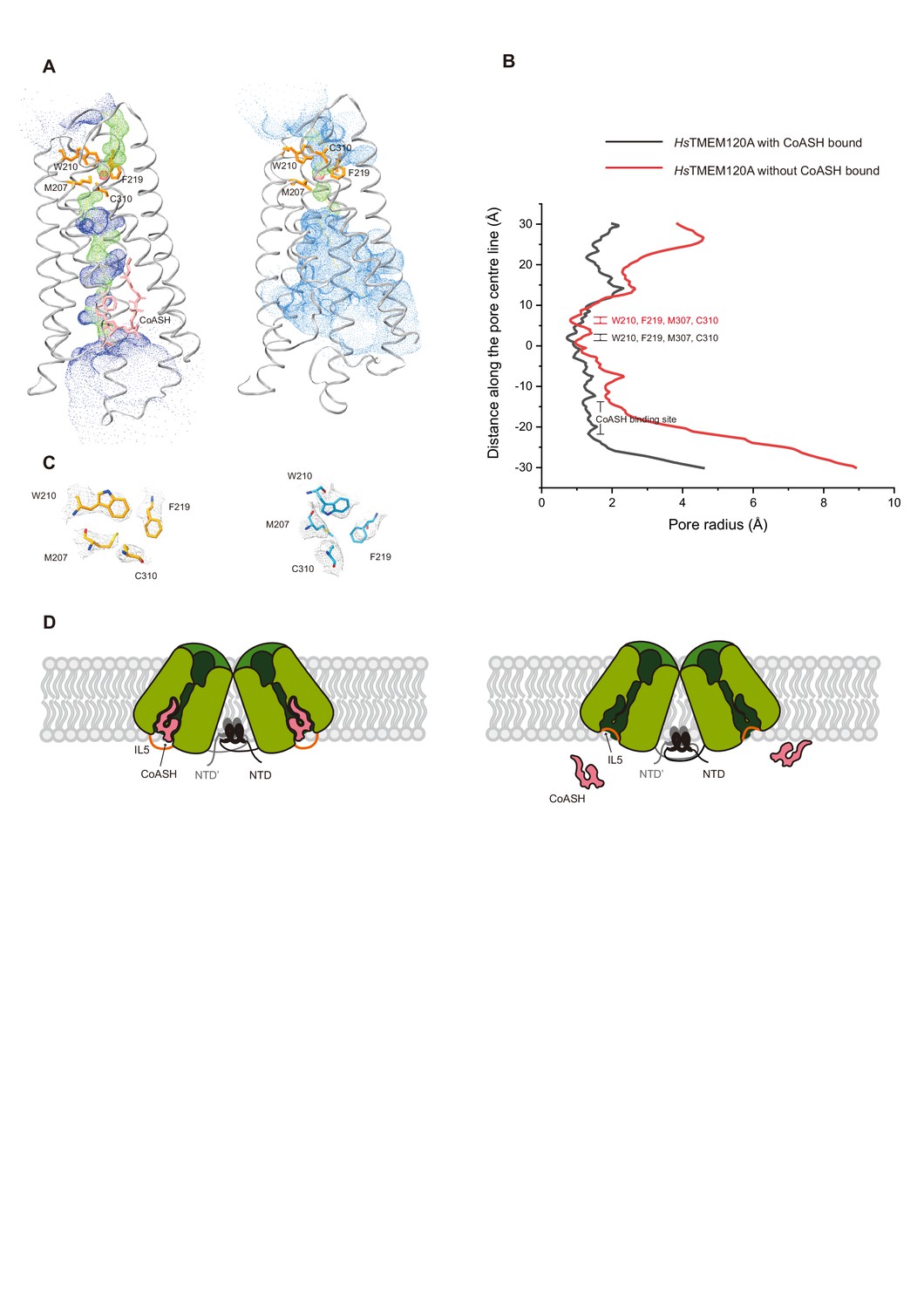
TMEM120A contains a specific coenzyme A-binding site and might not mediate poking- or stretch-induced channel activities in cells | eLife

IJMS | Free Full-Text | Computer Simulations and Network-Based Profiling of Binding and Allosteric Interactions of SARS-CoV-2 Spike Variant Complexes and the Host Receptor: Dissecting the Mechanistic Effects of the Delta and

Development of a coupled pile-to-pile interaction model for the dynamic analysis of pile groups subjected to vertical loads | SpringerLink

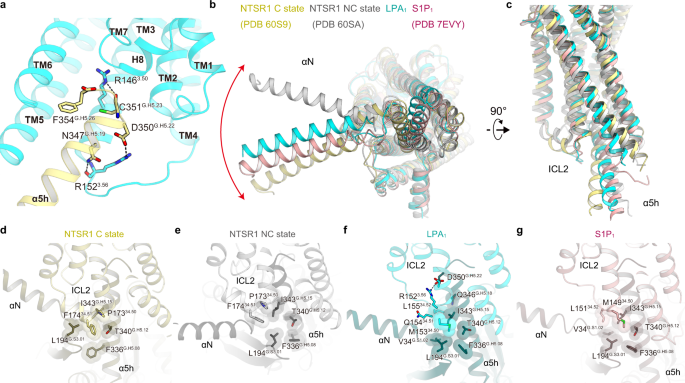
Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate | Nature Communications

Atomistic Simulations and Network-Based Energetic Profiling of Binding and Allostery in the SARS-CoV-2 Spike Omicron BA.1, BA.1.1, BA.2 and BA.3 Subvariant Complexes with the Host Receptor: Revealing Hidden Functional Roles of the

X-ray structure of fucose in the additional binding site IV of D-PLL... | Download Scientific Diagram

Elucidation of interactions regulating conformational stability and dynamics of SARS-CoV-2 S-protein - ScienceDirect

Dynamic Interactions of Fully Glycosylated SARS-CoV-2 Spike Protein with Various Antibodies | bioRxiv

Further thermo‐stabilization of thermophilic rhodopsin from Thermus thermophilus JL‐18 through engineering in extramembrane regions - Akiyama - 2021 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
Structure of the active Gi-coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist | Nature Communications
Integrating Conformational Dynamics and Perturbation-Based 2 Network Modeling for Mutational Profiling of Binding and Allostery

Dynamic Interactions of Fully Glycosylated SARS-CoV-2 Spike Protein with Various Antibodies | Journal of Chemical Theory and Computation
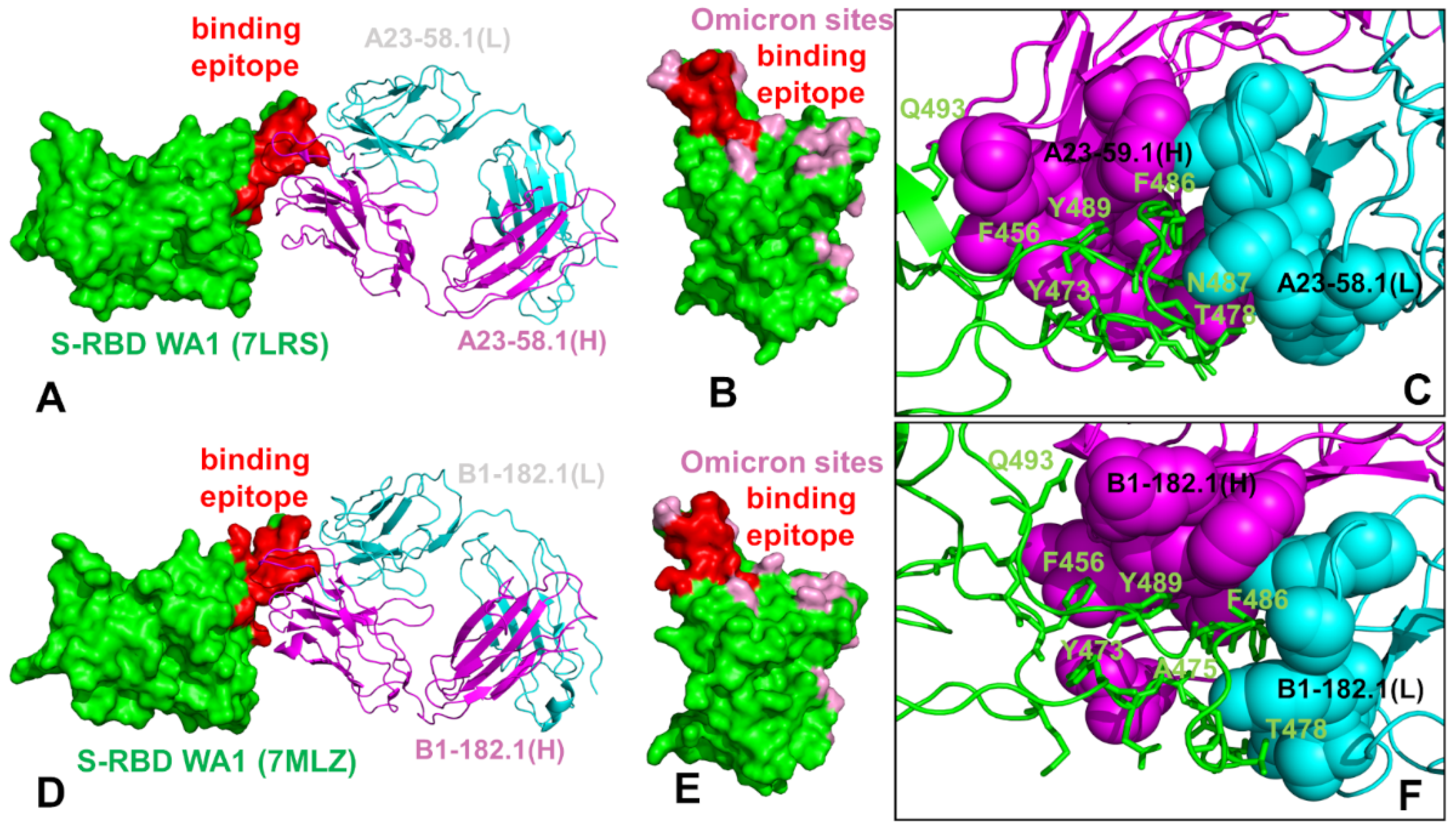
Biomolecules | Free Full-Text | Integrating Conformational Dynamics and Perturbation-Based Network Modeling for Mutational Profiling of Binding and Allostery in the SARS-CoV-2 Spike Variant Complexes with Antibodies: Balancing Local and Global Determinants

Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C6‐Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C6‐Methyltransferase - Tsutsumi - 2022 - Angewandte Chemie International Edition - Wiley Online ...

Accurate 3D Characterization of Catalytic Bodies Surface by Scanning Electron Microscopy - Gontard - 2019 - ChemCatChem - Wiley Online Library




